Progress
2016
Collection of contemporary samples
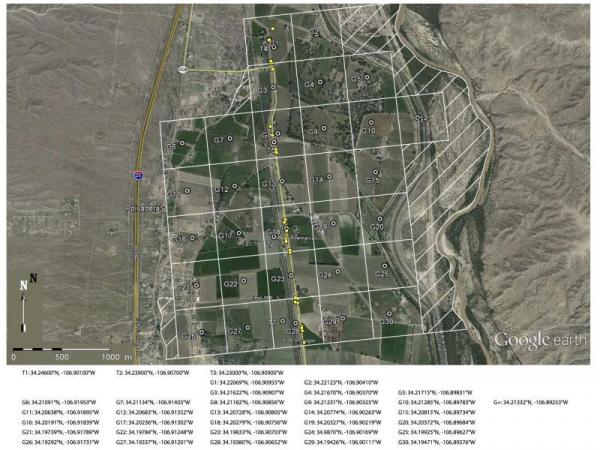
Three collecting trips were completed during 2015-2016. The first two were conducted to locate the current location of the advancing G. aurei populations. During the third trip, a large field party including Co-PIs Demastes, Spradling and Light, three students, and collaborators D. Hafner and M. Hafner extensively sampled the wave front using a GPS-coordinated collecting grid to allow for specific testing of geographic patterns of allelic distributions (se figure). Sampling also took place North of the San Acacia constriction to resample core populations as well as at the site of the wave front in 1991 to test genetic recovery hypothesis. 98 pocket gophers and their associated louse populations were collected during this trip representing all points on the grid overlay.
2016-2018
Genetic analysis
Microsatellite data was generated for both the historical and contemporary samples collected. A total of 67 louse populations were sampled with a target of 30 lice examined per population. This resulted in over 2020 chewing lice being analyzed and preserved as morphological vouchers. 15 polymorphic microsatellite loci were examined for each louse. Hypothesis testing will begin during the summer of 2017.
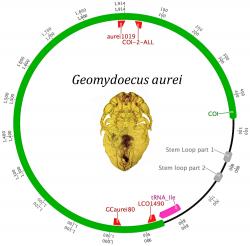
In an attempt to produce another measure of population genetic diversity to use in comparison with microsatellites, we studied the mitochondrial DNA (mtDNA) of these lice looking for hyper-variable regions that would be informative. During the course of this work, it was determined that the mtDNA of Geomydoecus aurei is fragmented into several minichromosomes, which is very unusual for most insects, but not for lice. Undergraduate researcher Lucas Pietan mapped the minicircle for the COI for G. aurei and for Thomydoecus minor (also found on the pocket gophers of San Acacia). Not only did this research demonstrate the presence of a fragmented mtDNA genome, but it also demonstrated the occurrence of heteroplasmy - the presence of multiple different copies of the mtDNA genome within an individual, a finding that we published in PLoS one (below).
 Pietan, Lucas L., Theresa A. Spradling, and James W. Demastes. (2016). The mitochondrial cytochrome oxidase subunit I gene occurs on a minichromosome with extensive heteroplasmy in two species of chewing lice, Geomydoecus aurei and Thomomydoecus minor. PloS one 11.9: e0162248.
Pietan, Lucas L., Theresa A. Spradling, and James W. Demastes. (2016). The mitochondrial cytochrome oxidase subunit I gene occurs on a minichromosome with extensive heteroplasmy in two species of chewing lice, Geomydoecus aurei and Thomomydoecus minor. PloS one 11.9: e0162248.
2018
In a truly collaborative effort, a suite of variable, reliable microsatellite loci were developed for the San Acacia Zone study:
2019
Two more publications are now out and/or accepted:
2019 J. W. Demastes, D. J. Hafner, M. S. Hafner, J. E. Light and T. A. Spradling. Loss of genetic diversity, recovery and allele surfing in a colonizing parasite, Geomydoecus aurei. https://doi.org/10.1111/mec.14997
2019 D. J. Hafner, M. S. Hafner, T. A. Spradling, J. E. Light, and J. W. Demastes. Temporal and spatial dynamics of competitive parapatry in chewing lice. Ecology and Evolution, Accepted: 11 March 2019.
